Microbes play a pivotal role in the food industry. Their metabolic activity through fermentation underlies the production of a large part of our diet, from fermented beverages like wine, beer and kombucha to fermented dairy products such as kefir, yoghurt and cheese.
Cheese fermentation involves complex interactions among several microbial species. Alongside traditional and modern cheesemaking processes, the wide diversity of microbes used in cheese fermentation results in a rich global cheese platter, placing cheese among the top foods in terms of its significant cultural dimensions and socioeconomic relevance.
While much is known about the effect of individual microbes on cheesemaking, less is understood about their interactions within the context of the microbial community.
To shed light on the molecular mechanisms underlying such complex interactions, a project named FoodTranscriptomics was initiated by Chr. Hansen, a Denmark-based global bioscience company. The multidisciplinary expertise among the research partners spanned microbiology, food science, biochemistry, bioinformatics and mathematical modelling.
A multi-year research project
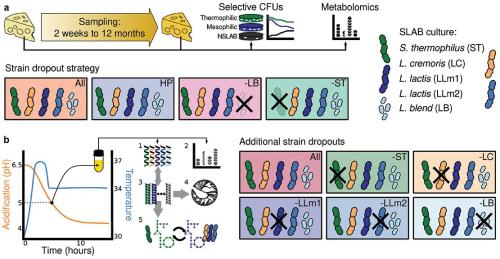
In cheesemaking, the initial step of milk fermentation is fairly well studied. This project instead focused on the long period of cheese ripening, which has a significant effect on the final flavour of the cheese. The main work started with a year-long experiment involving the production of cheddar cheese while varying the key members of a multi-strain commercial cheese starter culture comprising:
- Streptococcus thermophilus
- two major Lactococcus lactis strains
- one major L. cremoris strain
- a mixture of 21 L. cremoris and L. lactis strains (Fig. 1a).
The microbial dynamics were then investigated by selective plating and counting of bacterial colonies after 2 weeks and then after 3, 6, 9 and 12 months.
In addition, an array of targeted biochemical measurements were employed at the same time intervals to understand the chemical changes during cheese ripening. Advanced technologies, such as liquid and gas chromatography combined with mass spectrometry, were used to measure carbohydrates, organic acids, peptides, amino acids and volatile compounds (Fig. 1a).
To gain a deeper understanding of the molecular mechanisms underlying the microbial interactions in cheese fermentation, we devised a second experiment involving controlled milk fermentations where additional strains were removed from the starter culture and various samples were harvested at pH 5 (Fig. 1b). Here we took an integrative systems biology approach that combined genomics, genome-scale metabolic modelling, metatranscriptomics and exo-metabolomics.
Metabolic interactions between microbes shape the biochemical profile of cheese
The results of the year-long cheesemaking experiment indicated that S. thermophilus benefits the growth of the lactococci community. This was evidenced by the significantly fast decline in the Lactococcus population when S. thermophilus was removed from the culture. In addition, the metabolic activity of S. thermophilus had a significant impact on the chemical profile of the cheese from the early stages to the final stage of the ripening process. This was especially pronounced in the final peptide compositions we observed.
Digging deeper to identify the underlying mechanisms revealed that S. thermophilus, through its proteolytic enzymes, provides peptides and amino acids as nitrogen sources to Lactococcus. Metabolic modelling and metatranscriptomics analysis revealed that S. thermophilus is likely to excrete branched-chain amino acids as a result of its metabolic activity. When S. thermophilus was removed from the starter culture, the Lactococcus community exhibited a significant upregulation in nucleotide biosynthesis through glutamine, purine and pyrimidine biosynthesis pathways, which are essential for cell maintenance.
While our results suggested a collaborative interaction between the microbes, we also observed competition between L. lactis and L. cremoris for available nutrients in the milk, such as citrate. C4 aroma compounds, such as diacetyl and acetoin, are derivatives of citrate metabolism and lead to a buttery flavour. By investigating the transcriptome profile of the microbial community, we found that all Lactococcus strains are likely to take up citrate.
While L. lactis appears to be primarily directed towards the production of diacetyl and acetoin, affecting the cheese’s buttery traits, L. cremoris exhibits a transcriptionally active pathway that produces α-ketoglutarate. L. cremoris also possesses active genes associated with aconitate hydratase and isocitrate dehydrogenase. These are involved in intracellular metabolic conversions and are not as relevant for flavour.
When L. cremoris was removed, we detected four flavour compounds. These included 2,3-pentanedione (which gives the flavour of nuts, cream and butter), and heptanal and hexanal (which taste fruity and fatty). On the contrary, the presence of L. cremoris led to higher amounts of 2-methyl-3-thiolanone (which adds a meaty flavour) as well as the esters ethyl acetate and ethyl hexanoate (which add a fruity flavour).
Overall, these findings led us to hypothesise that competition between bacterial species induces changes in crucial flavour compounds. These include diacetyl, acetoin and other compounds with unknown biosynthetic reactions responsible for the production of 2,3-pentanedione, esters and more. These compounds are typically associated with fruity, creamy, buttery and nutty flavours.
Our results show how strain-specific metabolic interactions between microbes shape the biochemical profile of cheese, and provide targets towards the rational design and assembly of microbial communities with the aim of fine-tuning the flavour of cheese. More broadly, our study provides a blueprint for uncovering in situ interactions in complex food microbial ecosystems.
The future awaits
Since an actual cheddar cheesemaking process was conducted, our findings are directly applicable to commercial cheese production processes. One of the key findings is that realising the desired flavour of a cheese depends not only on individual microbial traits but also on various interactions among microbes, even at a strain level. Our findings thus open the door to investigate microbial interactions in a wide range of cheeses and other types of fermented foods, as the microbes used here – S. thermophilus, L. lactis and L. cremoris – are heavily used in the fermented food industry.
The results resonate outside the dairy industry. An example is the growing area of plant-based food fermentation, where the removal of off-flavours and toxic compounds is essential for a pleasant and safe final product.
We have also shown that varying the conditions can switch collaborative microbial interactions to competitive interactions, and vice versa. This has also been observed by our project partners in kefir fermentation (doi.org/10.1038/s41564-020-00816-5).
Therefore, we believe that the nature of microbial interactions in a community should always be investigated in the context of the environmental conditions applicable to the community. The ultimate objective would be to rationally design microbial communities for food fermentations based both on individual traits and the potential interactions of microbes to achieve the desired food characteristics. To augment this mechanistic approach, data-driven mathematical models using AI can be of great value in accounting for unknown mechanisms, and thus accelerate the development of “designer” microbial cultures for food fermentations.
Beyond the realm of cheese, the FoodTranscriptomics project also explored the impact of microbial biochemical activity on reducing the alcohol level in wine. That research has been published in Food Microbiology (doi.org/10.1016/j.fm.2022.104167).